Introduction
CRISPR-Cereal is a regulome and single nucleotide polymorphism (SNP) information based CRISPR guide RNA design tool for wheat, maize and rice. Four questions CRISPR-Cereal can help to answer, i.e, 1) The best candidate gRNAs with the highest prediction on-target scores in the requested region. 2) The number and efficiency of the off-target sites for candidate gRNAs in genome. 3) The chromatin environment in the target sites complementary to the candidate gRNAs. 4) The existed SNPs on the target sites.
Quick start
The workflow of CRISPR-Cereal is presented in Figure 1.
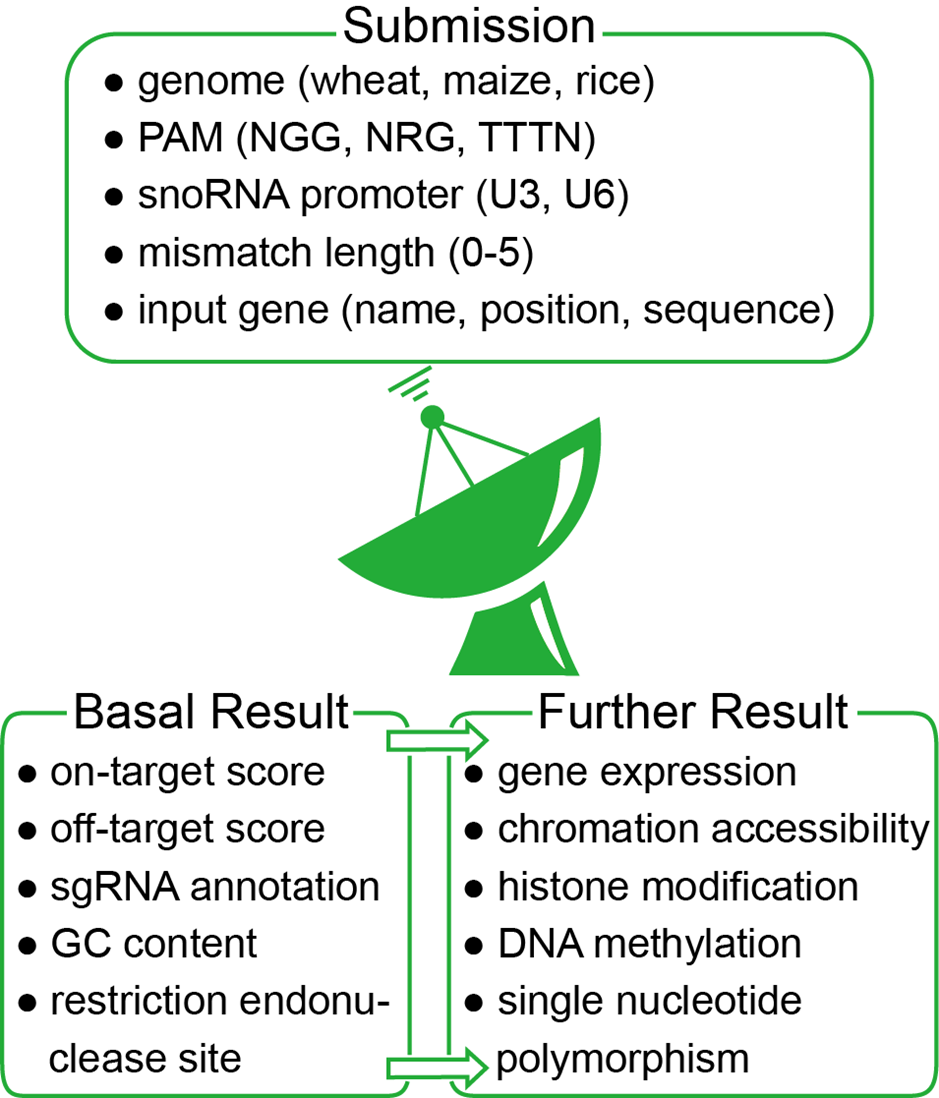
Firstly, you need to select a genome, choose the PAM and input the gene name, locus or sequence in the “Submit” page. The maximum mismatch length could as default when click the “Submit” button to start a job.
Next, the “Result” page would automatically appear after job submission. All the putative gRNAs are listed from high on-target score to low score with the information of sequences, GC contents, positions, proximate genes, location of gene structure elements and off-target amounts. Click the off-target number, the information of the top 20 off-target sites would be presented, including the sequences, mismatch number and bases from on-targets, positions, proximate genes, location of gene structure elements and the off-target scores. The sequences and information of the whole off-target sites could be downloaded when click the “download all offtarget” button located on the top left corner on the result page.
Following, click the position of each candidate sgRNA, and then the web browser would skip to an newly constructed Generic Genome Browser (GBrowse) web page1. The regulome and SNP information of each on-target site are showed.
Step by step manual
Step 1: Submission
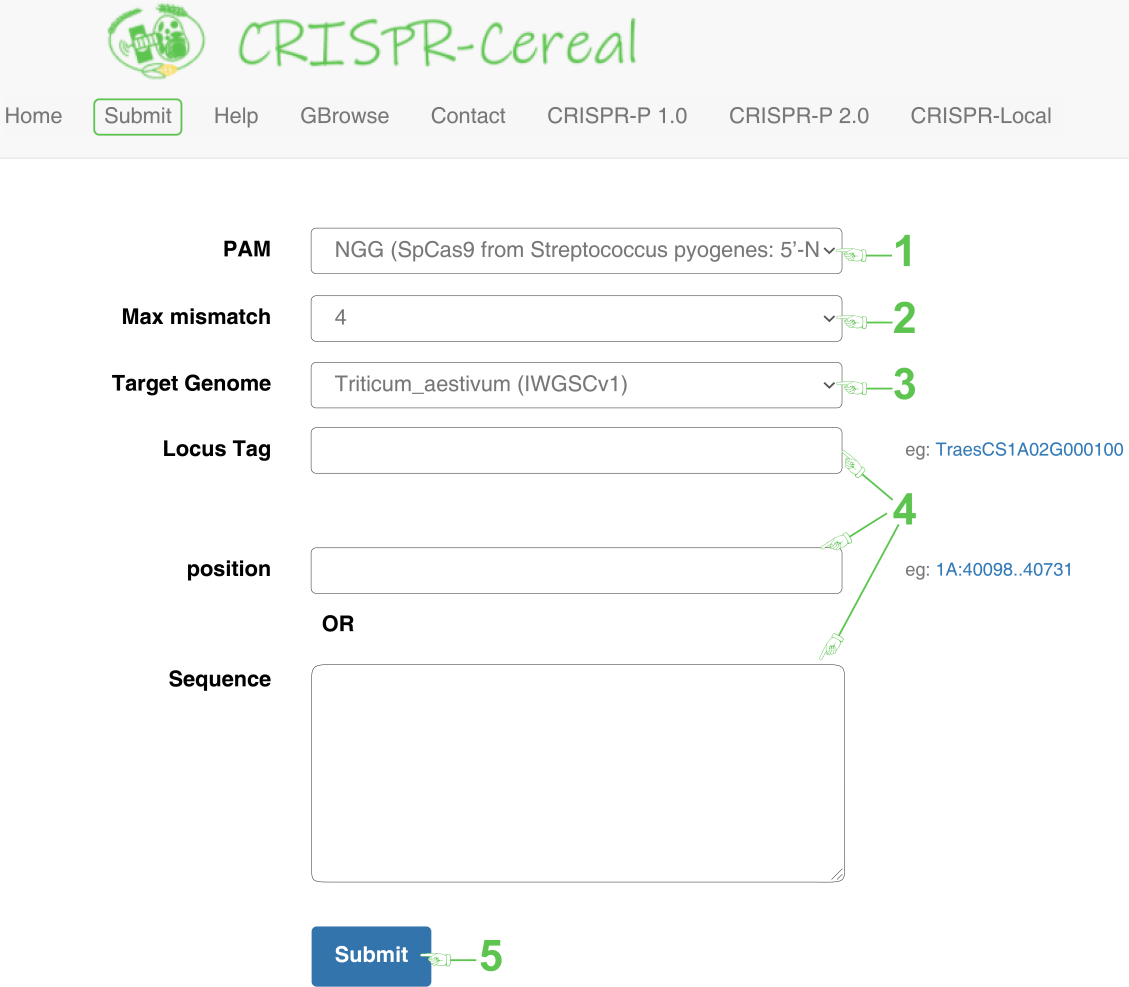
The “Submit” page would be accessible after clicking the “Submit” button on the menu bar (Figure 2).
1. Select a PAM (protospacer adjacent motifs), which is a short sequence immediately following the target sites and specific to each CRISPR-Cas subtype and bacteria. CRISPR-Cereal provides the most widely used type of PAM, including 5'-NGG-3' or 5'-NRG-3' (R = A, G) for Streptococcus pyogenes Cas9 and 5'-TTTN-3' for Acidaminococcus Cpf1 or Lachnospiraceae Cpf1.
2. Defined the max number of mismatch base pairs differ from on-target sequence. Once the max mismatch number has been set, CRISPR-Cereal would find the off-target sequences with mismatches no larger than the defined number. Note that the load time of the result page is proportional to the mismatch number. The mismatch number may be 0, 1, 2, 3, 4 or 5 and the default is 4.
3. Select the target genome, i.e, wheat, maize, indica rice or japonica rice. The genome version for wheat is IWGSC v1.1 downloaded from Ensembl Plants (ftp://ftp.ensemblgenomes.org/pub/plants/release-47/), maize is AGP v4.0 downloaded from Gramene (ftp://ftp.gramene.org/pub/gramene/release-62/gff3/zea_mays/), indica rice is MH63RS3 provided by Professor Chen (llchen@mail.hzau.edu.cn; http://rice.hzau.edu.cn/rice/) and japonica rice is IRGSP-1.0 downloaded from MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). All the versions are most updated or most widely used, especially the gap-free genome version, MH63RS3.
4. Input the information of the target gene. You could choose to input the Locus tag, position or sequence. The input locus tag of wheat should be TreaCS##02G######, maize is Zm00001d#####, indica rice cv MH63 is OsMH_##G####### and japonica rice cv Nipponbare is LOC_Os##g#####. The input gene position is in Chromosome:start..end format, and the input gene sequence is in fasta format with a start symbol of “>”.
5. Click "Submit" to start sgRNA design.
Step 2: Choose guide RNA preliminary
After job submission, CRISPR-Cereal would search the on-target and off-target sites genome wide. All the on-target sites would be listed and sorted by the on-target scores. Moreover, the sequences, GC contents, positions, proximate genes, located gene structures, on-target scores and the off-target numbers would also be provided on the basal result page(Figure 3A, 3B). After click the the off-target number, the top 20 off-target sites would be displayed with information including sequences, number of mismatch base pairs from on-target sites, positions, proximate genes, located gene structures and off-target scores (Figure 3C). All the off-target sites could be downloaded after click the “download all offtarget” button (Figure 3D).
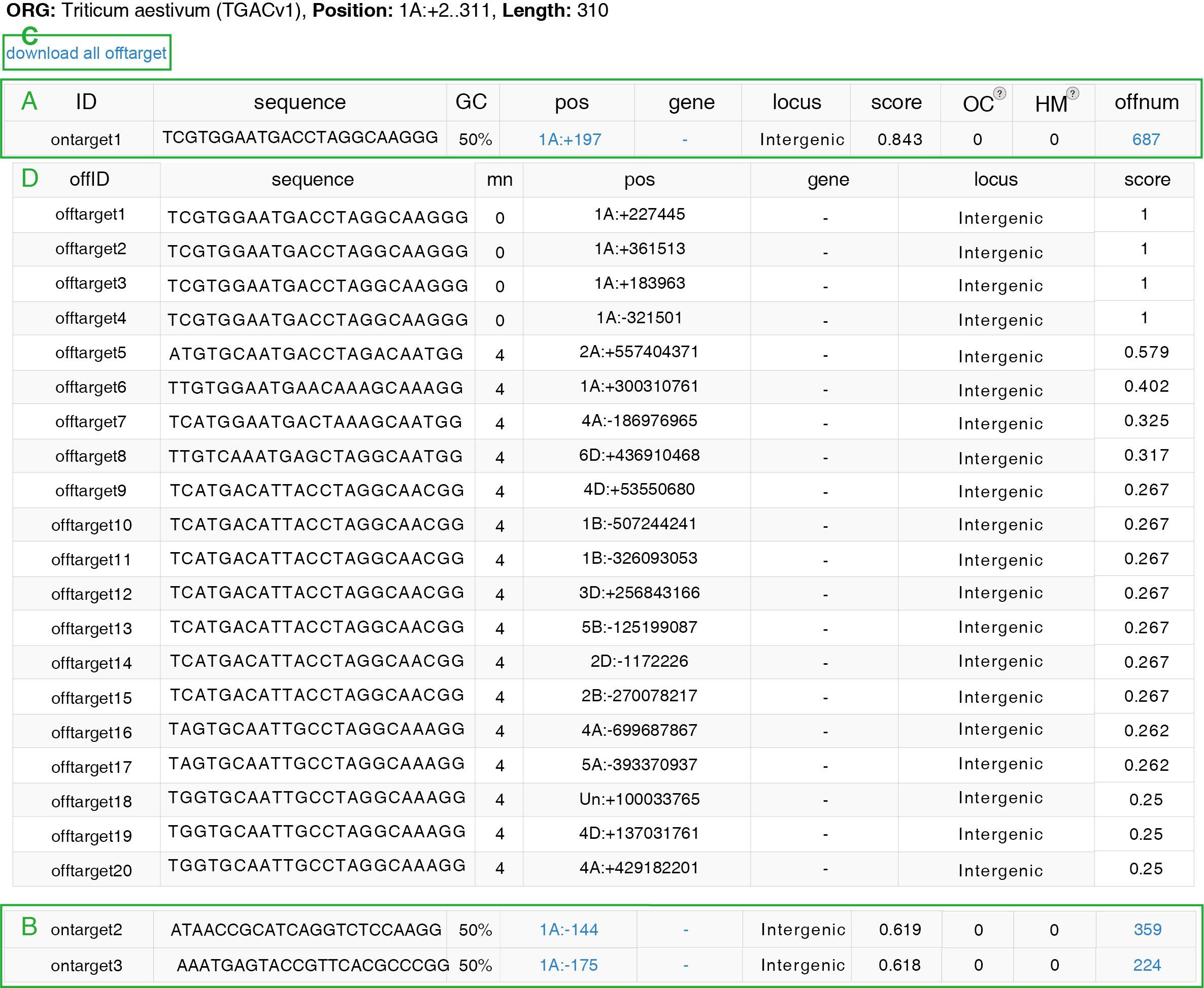
GC content: gRNAs with very low or high GC content tend to low efficiency3, 4. The GC-content of each sgRNA sequence is recommended to between 30% and 80%5. In addition, higher GC-content on the first 10 nt of the sgRNA (distal to PAM) was reported to achieve higher activity6.
On-target score: To reveal the underlaid rules of on-target efficiency in the CRISPR-Cas gene editing system. Doench and colleagues assessed the editing efficiency for a sgRNA pool in six endogenous mouse and three endogenous human genes3. As a result, they created a predictive model for the activity of sgRNA by using 1,841 gRNAs. CRISPR-Cereal applies this model to predict and score the on-target activities of gRNAs, and the sgRNA with the higher score is predicted to with higher editing efficiency.
Off-target score: The off-target is the obstacle for CRISPR-Cas gene editing. To reveal the sequence features for prediction of off-target activity, Doench and colleagues further used the sgRNA libraries designed according to the previously published rules, followed by large-scale assessment of the off-target activity. In the end, they developed a cutting frequency determination (CFD) score to predict the off-target efficiency7. The model Doench and colleagues built is robust and has been widely used. CRISPR-Cereal also uses this model to predict the off-target efficiency for each off-target site of each predicted gRNA. The gRNAs with low CFD scores for the off-target sites are recommend.
Step 3: Further selection of guide RNA
The chromatin environments and the SNPs of /on the target sites were reported to have influence on the editing efficiency of the CRISPR-Cas system8-10. Thus, CRISPR-Cereal collected high throughput sequencing data (Table 1) and visualized them using the GBrowse (Figure 4)8.
| Epigenetics | |
| wheat | ATAC-seq (GSE133885); DNaseI_seq (GSE121903); DNS-seq (GSE153289); ChIP-seq (GSE121903, SRP1262229); WGBS (SRP133674). |
| maize | ATAC-seq (GSE120304, GSE128434); ChIP-Seq (GSE120304, GSE128434); WGBS (GSE120304). |
| MH63 | GSE142570 (FAIRE-seq, eChIP-seq, WGBS) |
| transcriptome | |
| wheat | RNA-seq (SRP133837, SRP133885, ERP104790, GSE121903, GSE133884, DRP000768, ERP003465, ERP004505, ERP008767, ERP009837, ERP013829, ERP013983, ERP015130, ERP016738, SRP004884, SRP013449, SRP017303, SRP022869, SRP028357, SRP029372, SRP038912, SRP041017, SRP041022, SRP043554, SRP045409, SRP048912, SRP056412, SRP060670, SRP064598, SRP067916, SRP068156, SRP068165, SRP078208). |
| maize | RNA-seq (SRP162341, SRP112599). |
| MH63 | RNA-seq (GSE142570). |
| Nipponbare | RNA-seq (SRP188687, PRJNA597070, SRP062248, SRP131218, SRP068369, SRP009099, SRP056635, SRP065476, SRP1709322, SRP170613, SRP165785, SRP017256, SRP128142, SRP013556, SRP052310, SRP063555, SRP107231). |
| SNP | |
| wheat | http://wheat.cau.edu.cn/Wheat_SnpHub_Portal/snphub_wheat_NG19_2/ |
| maize | MaizeSNPDB (https://venyao.xyz/MaizeSNPDB/) |
| MH63 | In-house | Nipponbare | ftp://ftp.ensemblgenomes.org/pub/plants/release-48/variation/vcf/oryza_sativa/; RiceVarMap v2.0 (http://ricevarmap.ncpgr.cn/) |
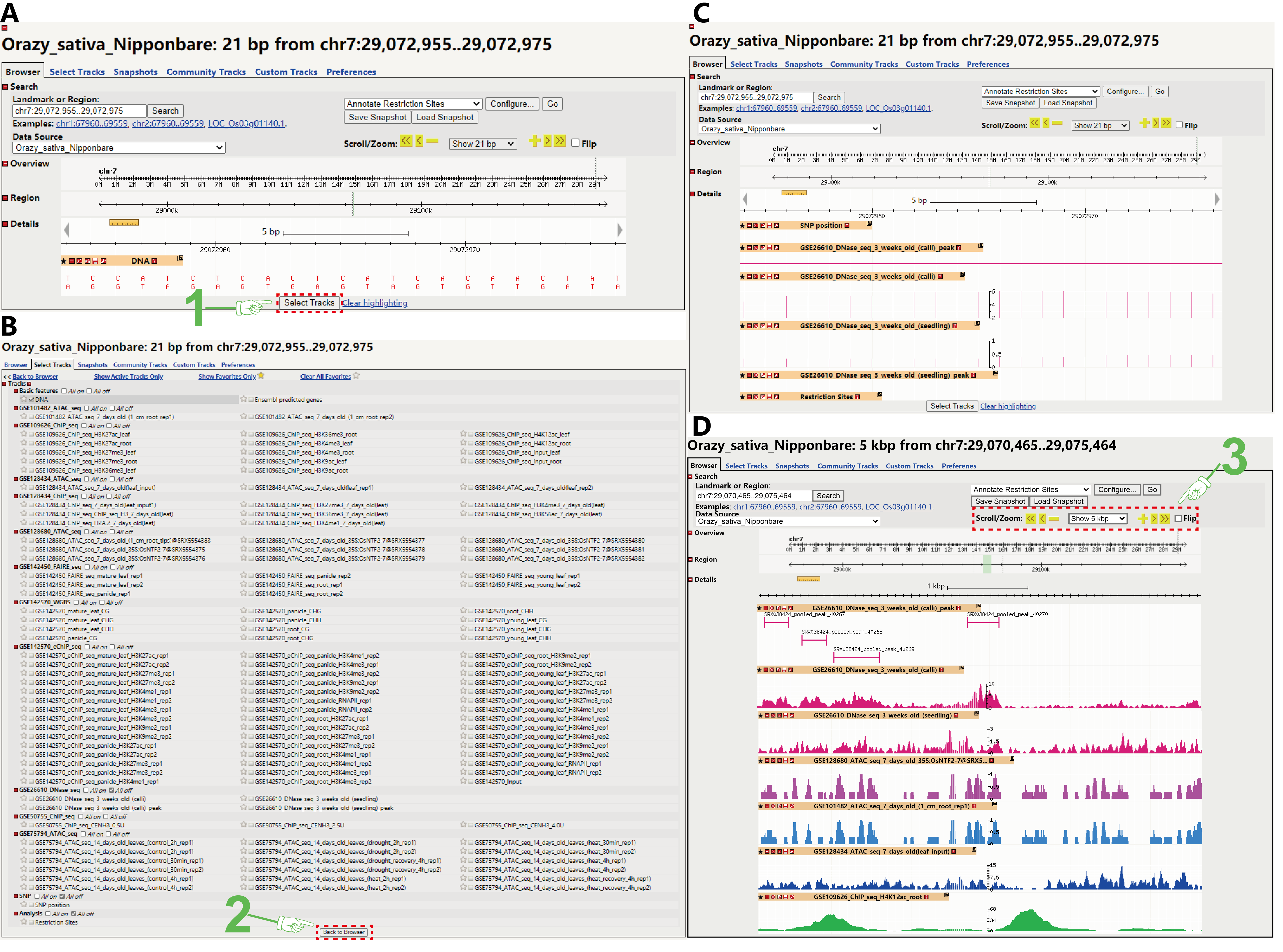
After click the on-target position on the basal result page (Figure 3A in blue), the web browser would automatically skip to the GBrowse web page. To get the regulome information of the gRNA, firstly, you need to select the concerned tracks which we uploaded in advance by click the “Select Tracks” button (Figure 4A). The tracks contain chromatin status information from ATAC-seq, DNaseI_seq, DNS-seq or FAIRE-seq, histone modification information from ChIP-seq or eChIP-seq, and DNA methylate modification information from whole genome bisulfite sequencing (WGBS) (Figure 4B). Also, the SNP information is added on the track (Figure 4B). You can trace back the data information by submitting the GSE IDs or SRR IDs we provided (track name) to the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database or the Sequence Read Archive (SRA, https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?) database, respectively.Besides, if you want to realize the putative restriction site located at the gRNAs, you can just tick the “Restriction Sites” track off.
Secondly, click the “Back to Browser” button, and then the chromatin states, epigenetic modifications and SNPs information of the 21 bp sgRNA targeted regions would be presented. The open chromatin and the epigenetic modification region is marked as anchored_arrows or with high scores (Figure 4C).
Thirdly, the default displayed region in the GBrowse is the 21 bp sgRNA target region, and only several separated bars in this region. While the open chromatin and the epigenetic marked regions are a set of peaks displayed on genome-wide. Thus, to better estimate the state of chromatin and epigenetic modification, you could zoom in a large range to see whether the 21 bp region located on a peak, and the gRNA target region would always at the midpoint among the zoomed region (Figure 4D). For more information about the data we provided, please see our publications. You can get more help to use the GBrowse by skimming the user manual on http://gmod.org/wiki/GBrowse_Tutorial.
To aid the gRNA design in particular cultivars, we also visualized the SNP information on the GBrowse. After click the SNP sites on GBrowse, the cultivars that contain the corresponding SNPs would be presented. For gRNA design towards particular cultivars, using gene Zm00001d023367 (10:4158140..4164969) as an example. If users want to design gRNA for W22 cultivar, the first step is to submit DNA sequence in FASTA format of the target gene to CRISPR-Cereal, the reference B73 genome will be selected to screen for on-targets (Figure 5A). Then, the on-targets information would be presented. Here, we choose one of the candidate gRNA (on-target8) and click the “position” (10: +4164938) (Figure 5B). Then, the page jumps to the GBrowse web page. The third step is to select the “SNP” track in GBrowse, and all the SNP located in the gRNA target region would be listed (Figure 5C). To see whether the W22 cultivar contains SNPs in the candidate gRNA or not, the forth step is to click the SNP and search for W22 in the “load_id” column (Figure 5D). If any SNP was found, just replace the base in the reference gRNA. Here base “C” in the 10:4164956 should be replaced to A for W22, and the gRNA sequence is GGAGACGAACAAAATCTGAAGGG. If no SNP is there, it indicates that the reference gRNA is suitable for W22, such as the first gRNA with highest on-target score.
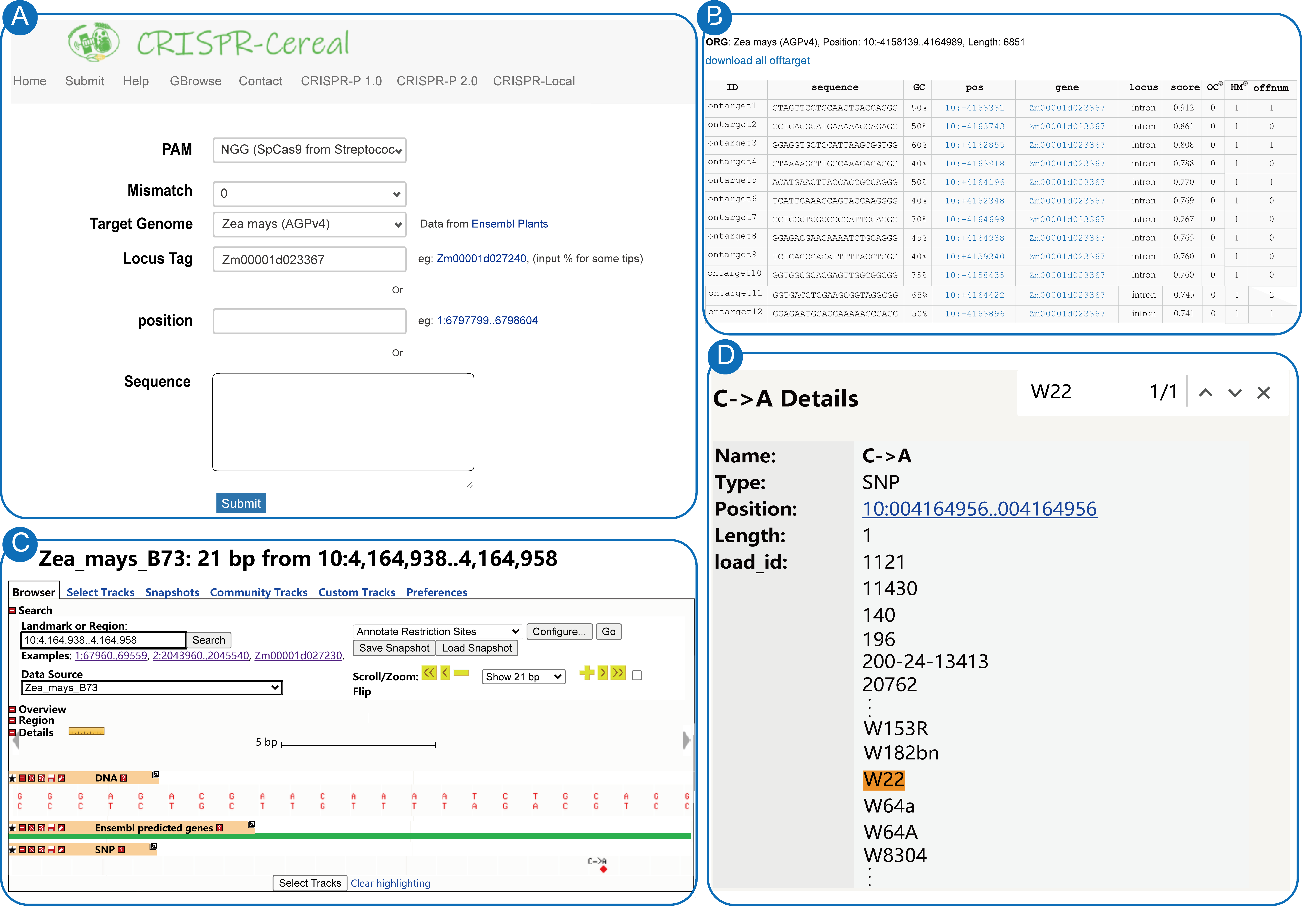
Note: The submit page (A) and basal result page (B) of gRNA design for gene Zm00001d023367 using CRISPR-Cereal. (C) The SNP information of the gRNA selected at the result page. (D) All the cultivars that show variations at the SNP site in (C), W22 is in orange after search using “Ctrl + F”.
Lastly, as the basal gene expression level is important for gene editing, we visualized the gene expression level and normalized the expression data in transcripts per million (TPM) on a new wed page. And after click the gRNAs proximate genes on the gRNA annotation table at the basal result page (Figure 3A-B), the gene expression information would be presented soon.
Additional information
To validate the authenticity of the off-targets detected by CRISPR-Cereal, we compared the off-target screening results obtained from CRISPR-Cereal and Cas-OFFinder (http://www.rgenome.net/cas-offinder/). As showed in Figure 6, the off-target outcomes among CRISPR-Cereal and Cas-OFFinder are highly uniformity. While when the mismatch set to 0, the Cas-OFFinder would contain gRNA sequence, and thus the off-targets number from Cas-Offinder is one more than CRISPR-Cereal. Moreover, the “N” bases arose from sequencing would recognized as a mismatch bases in Cas-OFFinder that caused the off-target number more than CRISPR-Cereal.

To confirm the relationship between editing efficiency and chromatin accessibility on the gRNAs target sites, we randomly collected the published data for 84 endogenous sites and checked the chromatin accessibility on the corresponding on-target sites in rice callus (Zhang et al., 2012). Please click here to download the data. The chromatin state score of each gRNA was estimated by the average reads coverage depth calculated by MASC2 in the gRNA target region. Namely, the DNase-seq narrow peak in wiggle format was firstly obtained through pipelines recommend by ENCODE. Then the average score of the bins in wiggle file intersected with each gRNA target region plus PAM was calculated via bedtools and shell-awk with the command: bedtools closest -D ref -t all -mdb all | awk '{sum[$2]+=$11;a[$2]++}END{for(c in sum){print(c,sum[c]/a[c])}}'. As expected, gRNAs targeting open chromatin regions result significantly higher editing efficiency than those against un-open regions (P=0.002), as reported in mammalian cells[11].
Reference
[1] Stein LD, Mungall C, Shu S, et al. The generic genome browser: a building block for a model organism system database. Genome Res, 2002,12(10):1599-1610.
[2] Song J, Xie W, Wang S, et al. Assembly and validation of two gap-free reference genomes for Xian/indica rice reveals insights into plant centromere architecture. bioRxiv 2020.12.24.424073; doi: https://doi.org/10.1101/2020.12.24.424073..
[3] Doench JG, Hartenian E, Graham DB, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol, 2014,32(12):1262-1267.
[4] Wang T, Wei JJ, Sabatini DM, et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science, 2014,343(6166):80-84.
[5] Liang G, Zhang H, Lou D, et al. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep, 2016,6:21451.
[6] Labuhn M, Adams FF, Ng M, et al. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res, 2018,46(3):1375-1385.
[7] Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 2016,34(2):184-191.
[8] Haeussler M, Schonig K, Eckert H, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol, 2016,17(1):148.
[9] Kuscu C, Arslan S, Singh R, et al. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol, 2014,32(7):677-683.
[10] Wu X, Scott D A, Kriz AJ, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol, 2014,32(7):670-676.
[11] Kim, H.K., Kim, Y., Lee, S., et al. SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance. Sci. Adv, 2019,5(11):eaax9249.