CRISPR-Local: a local single-guide RNA (sgRNA) design tool for non-reference plant genomes
CRISPR-derived editing system has been widely used for genome editing, and reaching a high-throughput level recently with the genome-wide mutant library construction and large-scale genetic screening. And it's thus highly critical to design reliable single guide RNA (sgRNA) for target gene locus in high-throughput way, and to take the variations of genetic backgroud into consideration, which is especially important for plant genome editing studies since the transgenic receptor lines used usually have no good genome sequences and mismatchs are very common compared with the reference genome.
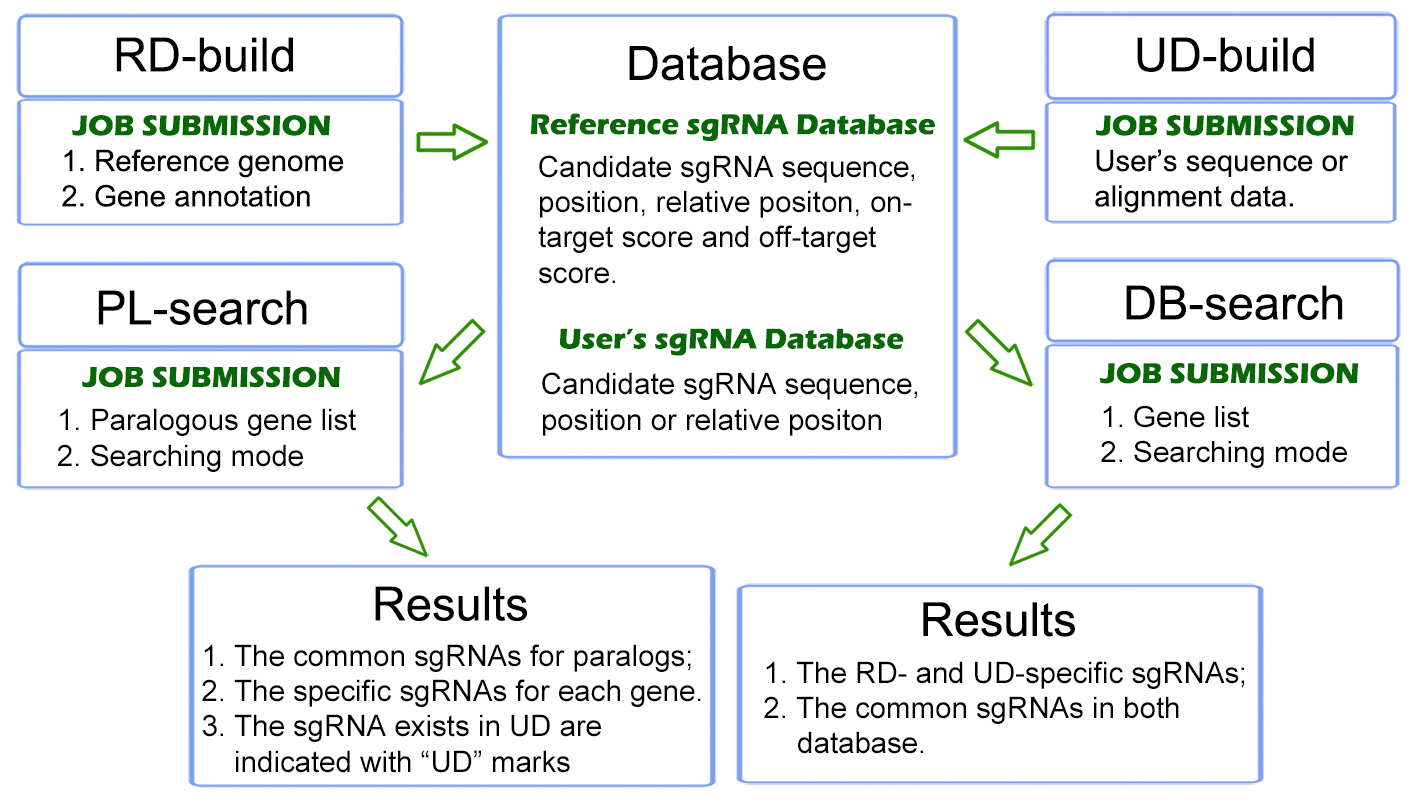
Here, we introduce CRISPR-Local for genome-wide sgRNA design with genotype-specific, by integrating the reference genome with various existing sequencing data on specific receptor lines. The major concepts of CRISPR-Local tool included two parts: the first is the "one-for-all" strategy to build whole genome sgRNA database with high efficiency, from which all possible sgRNAs for given reference (or user-defined genome) are generated and stored in local disk; the other is to retrieve (or de novo design) applicable hits through integrating data of whole genome sequencing (WGS), mRNA- sequencing, or known variants for specific transgenic receptors. A detailed description for CRISPR-Local could be available at "Help" page.
We have applied CRISPR-Local to a set of public plant genomes, which could be directly downloaded (see "download") for an easier following batch analysis, or can be quickly searched (see "DB-Search", reference genome based) for daily jobs with few loci concerned. Additionally, CRISPR-Local provides a function (see "PL-Search") to screen sgRNAs targeting multiple genes (usually paralogs) given an increasing requirements of editing multiple candidates at one event.
Reference
1. Jiamin Sun, Hao Liu, Jianxiao Liu, Shikun Cheng, Yong Peng, Qinghua Zhang, Jianbing Yan, Hai-Jun Liu*, Ling-Ling Chen*. CRISPR-Local: a local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics, bty970, https://doi.org/10.1093/bioinformatics/bty970
2. Hao Liu, Yuduan Ding, Yanqing Zhou, Wenqi Jin, Kabin Xie*, Ling-Ling Chen*. CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants. Mol Plant, 2017, 10(3): 530-532. doi: 10.1016/j.molp.2017.01.003 .
