CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants
if you have any problems or want any plant genome to be added to CRISPR-P 2.0, please visit this github repository(CRISPR-P 2.0) and make an issue.
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is an acronym for DNA loci that contain multiple, short, direct repetitions of base sequences. Each repetition contains a series of base pairs followed by the same or a similar series in reverse and then by 30 or so base pairs known as "spacer DNA". CRISPRs are found in the genomes of approximately 40% of sequenced bacteria and 90% of sequenced archaea. CRISPRs are often associated with cas genes which code for proteins that perform various functions related to CRISPRs. The CRISPR-Cas system functions as a prokaryotic immune system, in that it confers resistance to exogenous genetic elements such as plasmids and phages and provides a form of acquired immunity. CRISPR spacers recognize and silence exogenous genetic elements in a manner analogous to RNAi in eukaryotic organisms.
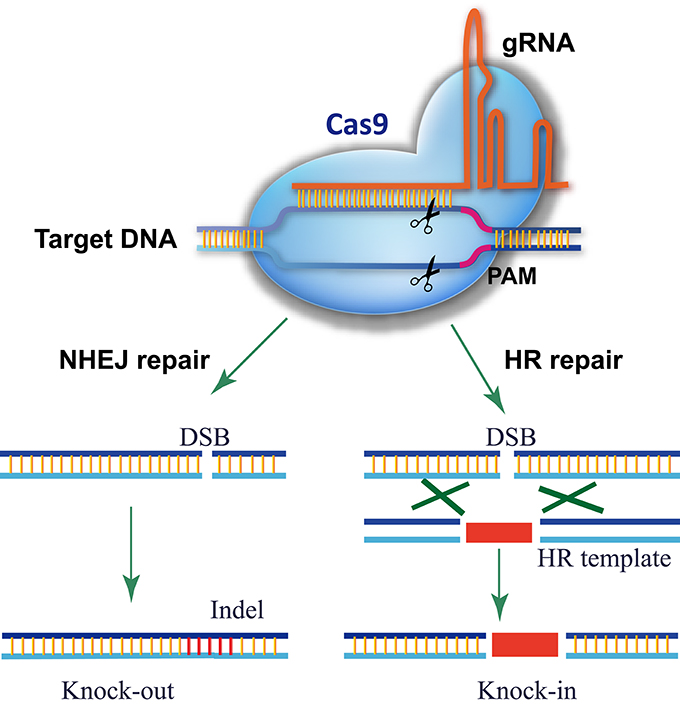
CRISPR-P 2.0 design tool based on we previously work of CRISPR-P (Lei et al., 2014), which is one of the most popular tools for sgRNA design in plants. As CRISPR/Cas9 technology has evolved rapidly in the past two years, an updated platform, CRISPR-P 2.0 provides web service for computer-aided design of highly efficient sgRNA with minimal off-target effects. It has functions as follow:
- It supports sgRNA design for 49 plant genomes. SgRNAs for 49 plant species with the latest version of genome and annotation are provided.
- Scoring system of sgRNA for on-target efficiency and off-target potential are improved. CRISPR-P 2.0 uses a modified scoring system to rate the off-targeting potential and on-targeting efficiency of sgRNAs for Streptococcus pyogenes Cas9 which is the widest used CRISPR-Cas9 system. The scoring system in CRISPR-P 2.0 is based on the up-to-date knowledge about SpCas9 genome editing.
- It Supports various CRISPR-Cas systems. It supports to design guide sequences for various CRISPR-Cas systems like Cpf1 (Zetsche et al., 2015) and various Cas9 endonucleases.
- A comprehensive analysis of the guide sequence is provided, including: GC content, restriction endonuclease site, microhomology sequence flanking the targeting site (microhomology score), and the secondary structure of sgRNA.
- Identification of sgRNA from custom sequences is also provided. If user’s genome/sequence is not listed in the selectable genomes, it allows users to upload custom sequences and identify sgRNAs.
Visit the old version of CRISPR-P
Reference
1. Yang Lei, Li Lu, Hai-Yang Liu, Sen L, Feng Xing, Ling-Ling Chen*. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant, 2014, 7(9): 1494-1496. doi: 10.1093/mp/ssu044.
2. Yuduan Ding, Hong Li, Ling-Ling Chen*, Kabin Xie*. Recent advances in genome editing using CRISPR/Cas9. Front Plant Sci., 2016, 7: 703.PMC4877526.
3. Hao Liu, Yuduan Ding, Yanqing Zhou, Wenqi Jin, Kabin Xie*, Ling-Ling Chen*. CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants. Mol Plant, 2017, 10(3): 530-532. doi: 10.1016/j.molp.2017.01.003 .
4. Jiamin Sun, Hao Liu, Jianxiao Liu, Shikun Cheng, Yong Peng, Qinghua Zhang, Jianbing Yan, Hai-Jun Liu*, Ling-Ling Chen*. CRISPR-Local: a local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics, bty970, https://doi.org/10.1093/bioinformatics/bty970
5. Chao He,Hao Liu,Dijun Chen,Wen-Zhao Xie,Mengxin Wang,Yuqi Li,Xin Gong,Wenhao Yan*,Ling-Ling Chen*. CRISPR-Cereal: a guide RNA design tool integrating regulome and genomic variation for wheat, maize and rice. Plant Biotechnol J. 2021 Nov;19(11):2141-2143. doi: 10.1111/pbi.13675
