Document
1. Background
The CRISPR/Cas9 technology is a modern, fashionable method in plant research. Immediately after its early use to edit the genomes of animals and bacteria, its efficacy was demonstrated in the model plant systems of Arabidopsis, rice, sorghum, and tobacco. Nowadays, this technology is broadly used in different plant species, and dozens of CRISPR/Cas9 vectors are available in the public plasmid repository of Addgene. The CRISPR/Cas9 pioneers have improved this system into a flexible and powerful platform for genome engineering. In this review, we briefly summarize the recent advances of CRISPR/Cas9 technology and its impact for plant genome engineering. More information could access in the review article (Ding et al., 2016): "Recent Advances in Genome Editing Using CRISPR/Cas9" in Front. Plant Sci.
We collected some of the key events in the development of CRISPR, click to the details.
The major concern about this technology is that Cas9 has off-target effects. Recent studies also showed that different guide sequences in sgRNAs have variable efficiency in genome editing (Doench et al., 2016; Fu et al., 2014; Liang et al., 2016). Therefore, the choice of targeting sites (same as the guide sequence of sgRNA) is the critical step in CRISPR-Cas9 technology.
2. Quick Guide for CRISPR-P 2.0 Users
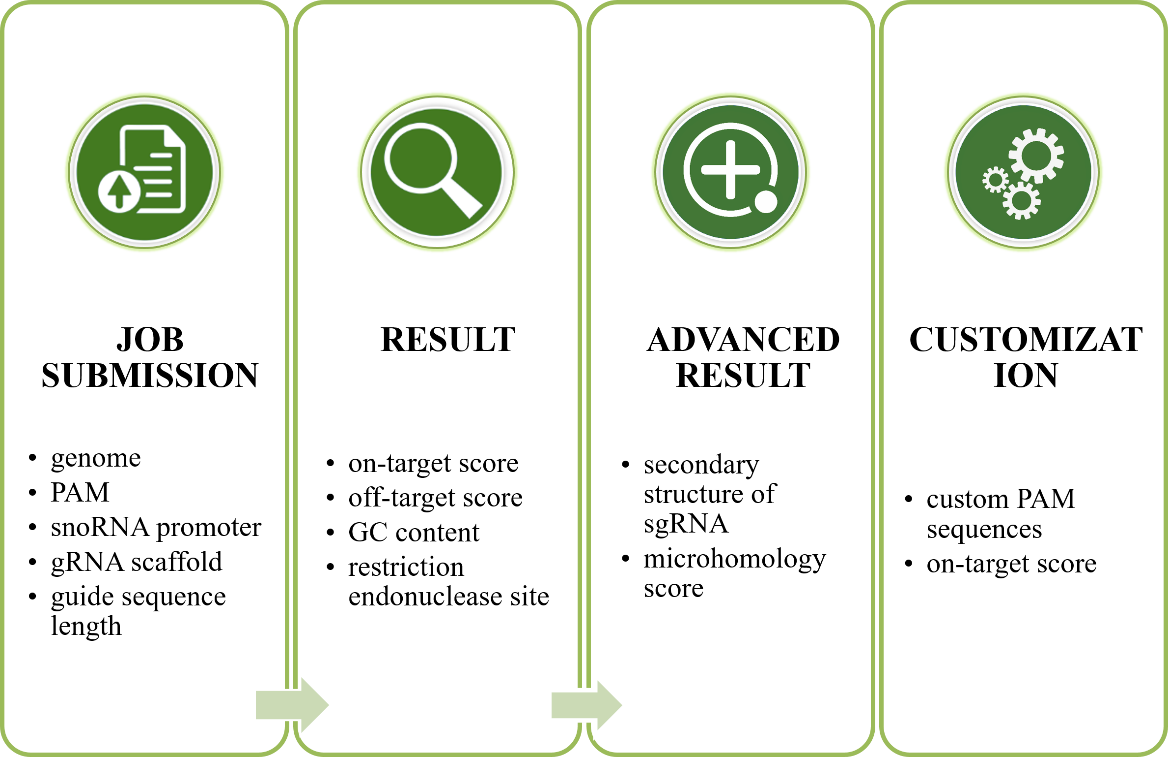
The following list provides a swift overview of the steps involved in sgRNA design (Figure 1). Note: Please read the step by step guide to prevent frustration and to ensure optimal results.
- Please access "Submit" page to start a job. Select a genome you are studying, input gene locus, chromosome position, or DNA sequence in FASTA format. Other parameters such as PAM, snoRNA promoter, RNA scaffold, guide sequence (spacer) length are also need be defined or with default arguments. Then, click "Submit" button. If your sequence could not be found in the blast search of your selected genome, you can jump to 4
- A few seconds after submission, the web browser would automatically enter the Result page of sgRNA design. Firstly, the target sequence is mapped to its genome. All possible sgRNA are screened out and showed in graphic genome model and information about potential on-target and off-target sits are displayed, including: on-target score, off-target score, GC content, restriction endonuclease site, etc.
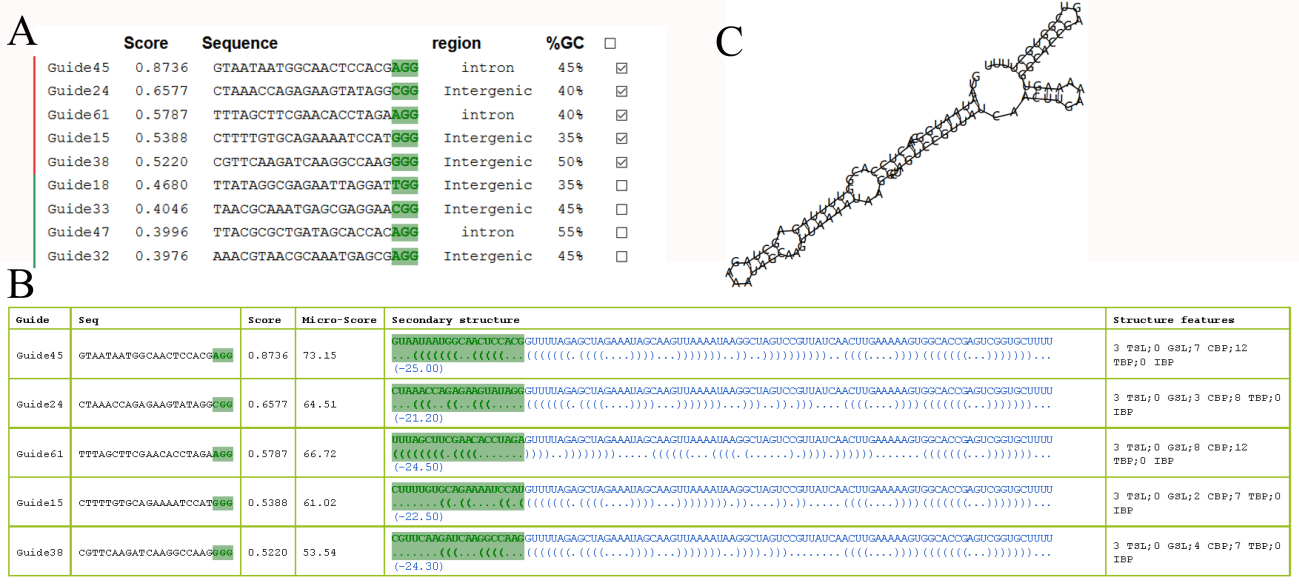
- Clicking "advance" button to enter the "Advanced result page of sgRNA design". It provided secondary structure of sgRNA and microhomology score for processes a further chose of efficient sgRNA. Designing sgRNAs according to your needs.
- User can identify on-target score from custom sgRNA sequence in "Design" page
3. Optimal sgRNA Design Step by Step in a Preset Genome
This section describes how to use the Optimized CRISPR-Plant 2.0 Design tool step by step in preset 49 plants. The following displays a detailed procedure for optimal sgRNA design.
- 3.1 Step 1 -Submit a job
- PAM
- snoRNA promoter
- RNA scaffold
- guide sequence length
- target genome
- sequence
- 3.2 Step 2 -Design optimized gRNA
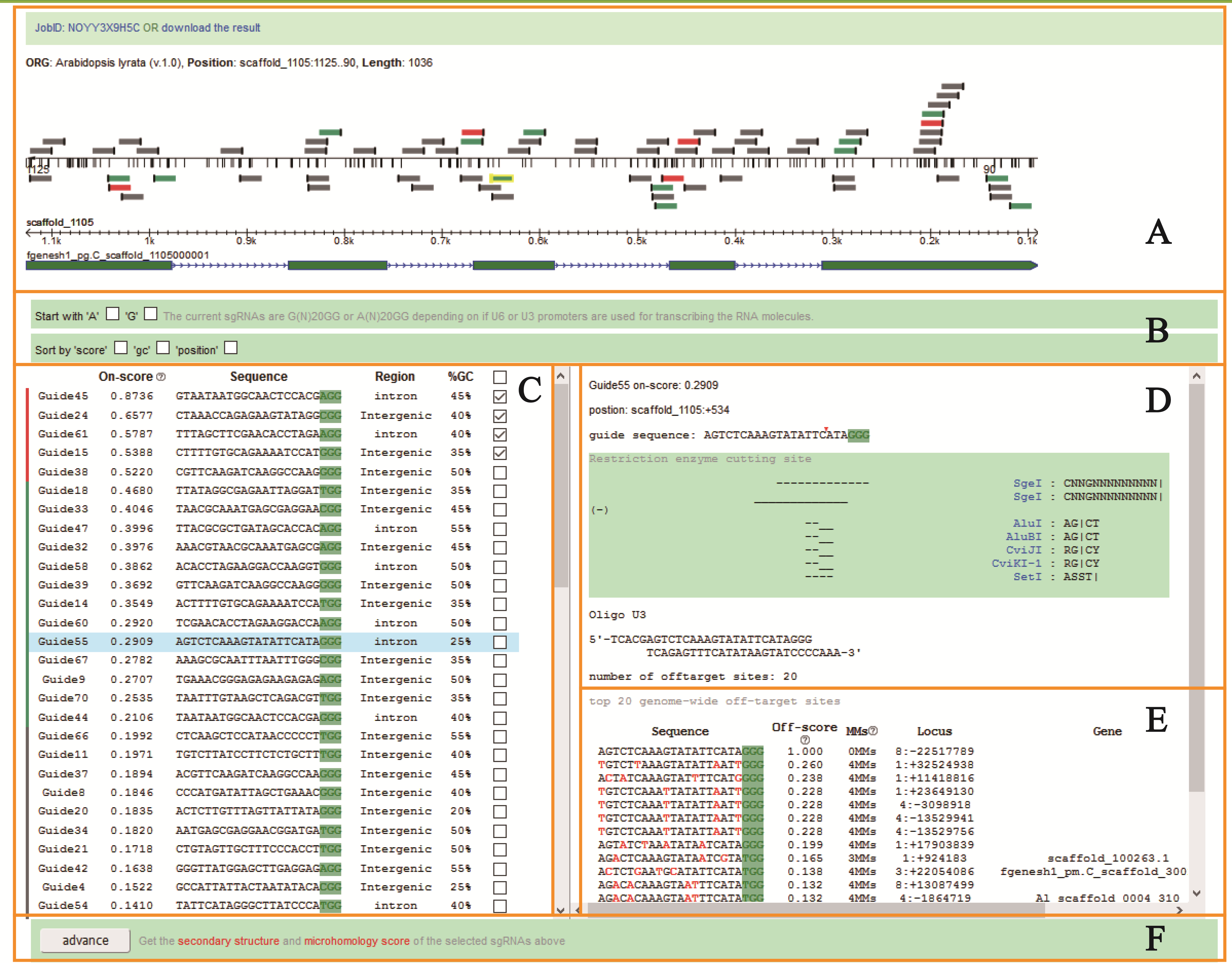
- The target sequence is mapped to its genome, and all possible sgRNA are screened out and showed in graphic genome model ( Figure 3A).
- On-target score: In order to access the on-target efficiency of sgRNA, we employed a scoreing module to evaluate the sgRNA provided by users in CRISPR-P 2.0. There were few known rules governing on-target efficiency of CRISPR/Cas9 system when CRISPR-P was developed. In recent years, Doench and colleagues discovered sequence features of sgRNAs that can improve on-target efficiency, and constructed a predictive model to design highly active sgRNAs, which includes the base distribution of 4 nt upstream of the sgRNA target site, the 20 nt of sgRNA complementarity, the PAM and 3 nt downstream of the sgRNA target sequence (Doench et al., 2014). A scoring module in CRISPR-P 2.0 was designed to access the on-target efficiency of sgRNA. It can identify potential sgRNAs and calculate their efficiency, and the prediction result is listed and scored ( Figure 3C). We colored in graphic genome model (Figure 3A): the presumably best ones in red (score > 0.50), followed by intermediate ones in green (0.20 < score < 0.50), and the remaining ones in grey at the bottom of the list. In additional, On-target Efficiency Score only support gRNAs of with 5'-NGG-3' PAM in Streptococcus pyogenes Cas9.
- Off-target score: off-target score module was designed based on the improved model of off-target according to their interference potential. Doench and his colleagues also profiled the off-target activity of thousands of sgRNAs and developed cutting frequency determination (CFD) score to predict off-target sites (Doench et al., 2016). We employed their models to predict the on-target and off-target activities of sgRNAs for CRISPR/Cas9 system in CRISPR-P 2.0. Every sgRNA sequences will be scanned for possible off-target matches throughout the selected genome. Then, top 20 off-target scored genome locus and its’ mismatches (MMs) to target sequence are listed for every sgRNA (the mismatch sites are highlighted with red color) (Figure 3E).
- GC content: It is reported that GC content of sgRNA is important for the efficiency of CRISPR/Cas9 system (Ren et al., 2014), and 97% of sgRNAs have a GC Content between 30% and 80% (Liang et al., 2016). For that reason, GC content of a sgRNA is also provided in CRISPR-P 2.0.
- Results content can download by click link "download the result" (Figure 3A) and view off-line.
- Preliminary screening of sgRNA. User can preliminary choose among the possible sgRNA according to its On-target/ off-target scores, location (gene exon, intron ,UTR or inttergenic), preferred restriction endonuclease site or CG content (30% - 80% CG content is recommended).
- 3.3 Step 3 - Advanced selection of sgRNA
- Microhomology Score: Bae and colleagues found that 39.6% of all the mutations induced by Cas9 RNA-guided engineered nucleases (RGENs) were associated with microhomology of 2-8 bases, then they developed a scoring system to estimate the frequency of microhomology-associated deletions at nuclease target sites (Bae et al., 2014). The scoring system of microhomology was embedded into CRISPR-P 2.0
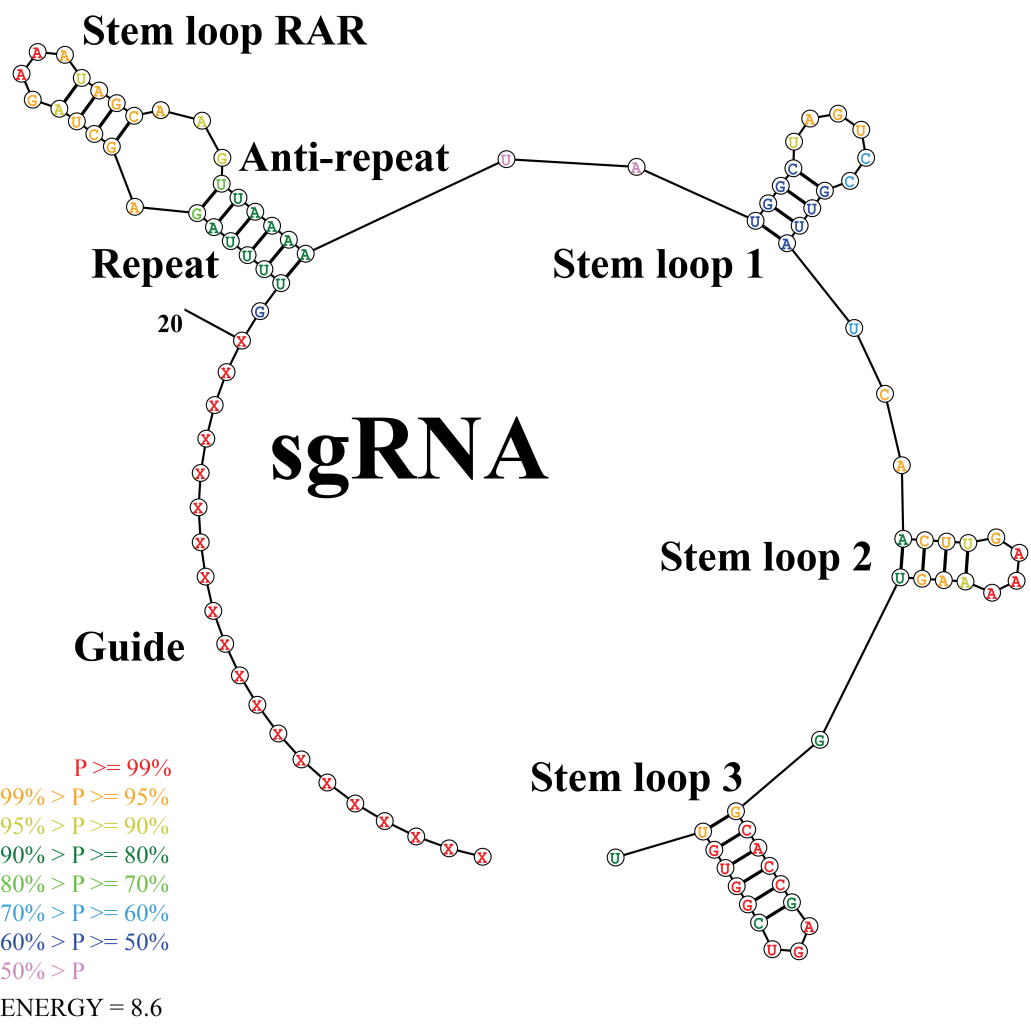
- Secondary structure of sgRNA: Researches (Fu et al., 2014; Liang et al., 2016) suggested that sgRNA functions by the interaction of its secondary structure with the Cas9 protein in vivo, the secondary structure of sgRNA may interfere with the editing efficiency and established links between secondary structure and editing efficiency of sgRNAs (Figure 4):
- The sgRNA contains crRNA- and tracrRNA-derived sequences connected by an artificial tetraloop. The crRNA sequence consists of guide (20nt) (also referred to spacer) and repeat (12nt) region, whereas the sgRNA sequence consists of anti-repeat (14nt) and three sgRNA stem loops.
- 97% of sgRNAs have a GC content between 30% and 80%.
- The repeat and anti-repeat region (stem loop RAR) triggers precursor CRISPR RNA (pre-crRNA) processing by the enzyme RNase III and subsequently activates crRNA-guided DNA cleavage by Cas9. It revealed that stem loop 1 is crucial for the function of Cas9-sgRNA-DNA complex.
- The stem loop 2 and 3 promote the stable complex formation. It implys that 3 stem loop structures are crucial for genome editing.
- Most of guide sequences in stable complex have characteristics that no more than 12 bases paring with the other bases of sgRNA, no more than 7 consecutive base pairs (CBPs), and highest internal base pairs (IBPs) number is no more than 6.
- Mark the preliminary chosen candidate sgRNA in result page (Figure 3A).Then, click "advance" button.
- The secondary structure and microhomology score of the selected sgRNAs are list as Figure 5 B-C.
- Micro-Score: the sum of all patten scores accroding to the microhomology size and the deletion length.
- Structure features: TSL: stem loop in total of the sgRNA; GSL: Stem loop in the guide sequence (20 nt); CBP: consecutive base pairs between guide sequence and the other sequence; TBP: total base pairs between guide sequence an the other sequence; IBP: internal base pairs in the guide sequence.
- Further selection of sgRNA could based on recommended criteria for selection of efficient sgRNAs as followed:
- G/C content between 30% and 80%;
- intact secondary structures: stem loop RAR, stem loop 1 and stem loop 2 (except for stem loop 1);
- no more than 12 TBPs;
- and no more than 7 CBPs;
- no more than 6 IBPs.
Please access "Submit" page to start a job (Figure 2). Optimized CRISPR-P 2.0 design tool supports preset 49 plant species genomes. To start a job, users could select one Target Genome and then submits a gene locus, chromosome position or DNA sequence for search. There are some other Parameters lists as follow:
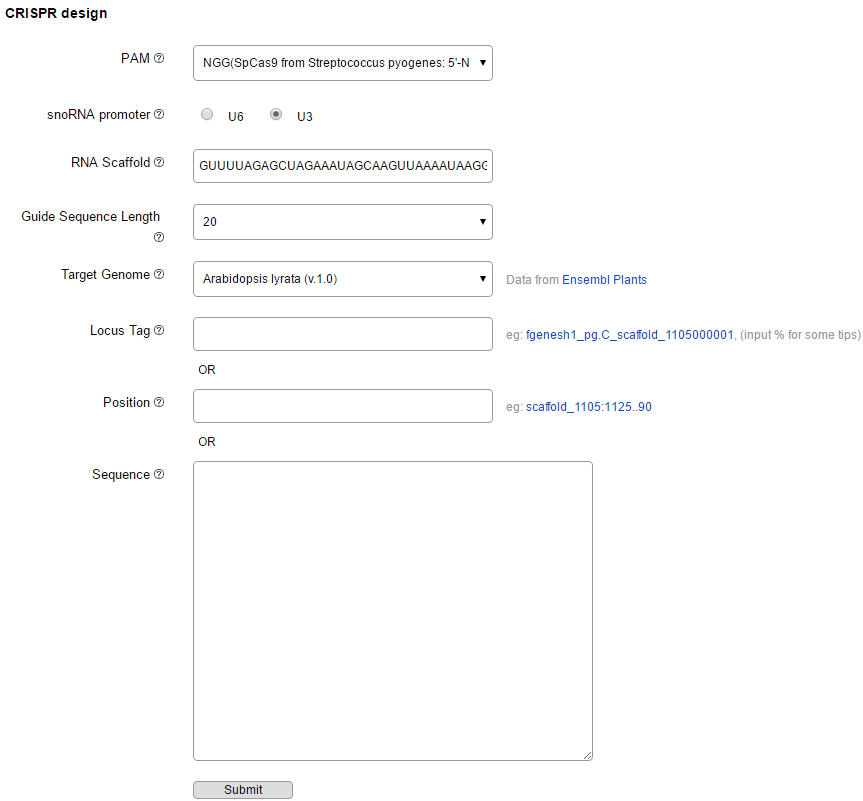
PAM is a DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease in the CRISPR bacterial adaptive immune system. The PAM sequence is absolutely necessary for target binding and the exact sequence is dependent upon the species. The Streptococcus pyogenes Cas9 (5'-NGG-3') is currently the most widely used in genome engineering. The additional species of Cas9 and corresponding PAM sequences are also provided in CRISPR-P 2.0, click to see the them
SgRNA have been expressed using plant RNA polymerase III promoters, such as U6 and U3 (Belhaj et al., 2013), U6 and U3 (default U3) are provided for optional promoters design in plant.
The RNA Scaffold can be customized and default as followed:
GUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUU.
The guide sequence (spacer) length can affect target efficiency and off-target potential. It is reported that truncated sgRNA which has targeting sequence is less than 20, could decrease undesired mutagenesis at some off-target sites (Fu et al., 2014).
Users can select different length of guide sequence (in the range of 15 - 22 bp). The spacer of sgRNA default contains 20 nucleotides which are complementary to a target DNA sequence.
There are preset 49 plant genomes provide in CRISPR-P 2.0. We will continuously update supported genomes once the high quality genomic sequence is published, click to the preset genomes.
The input DNA sequence in FASTA format is aligned to the target plant genome by using BLASTN, the best BLASTN hit with the same length, no mismatch and no gap is selected. Sequence in the range of 30 ~ 5,000 nt is suggested to input. If the input sequence is not in the BLASTN results of the target genome, the task will be stopped.
Click "Submit" button, the calculating will be very fast and longer sequence will need more times. The target sequence is mapped to its genome, and information about potential on-target and off-target sits are displayed in a result page including: on-target score, off-target score, GC content, restriction endonuclease site, etc. ( Figure 3)
User can process a further chose of efficient sgRNA in this step based on secondary structure and microhomology score (micro-score)
Introduction of microhomology score and secondary structure of sgRNA
The process of advanced selection of sgRNA as follow:
4. Identifying On-target Efficiency Score from Custom sequence
User can identify On-target Efficiency Score from custom sgRNA sequences in a standalone sgRNA design module. This module has two basic functions:
- Estimating On-target Efficiency Score from custom sgRNAs;
- Identifying sgRNAs from custom long squences and then estimate thise sgRNAs with on-target score;
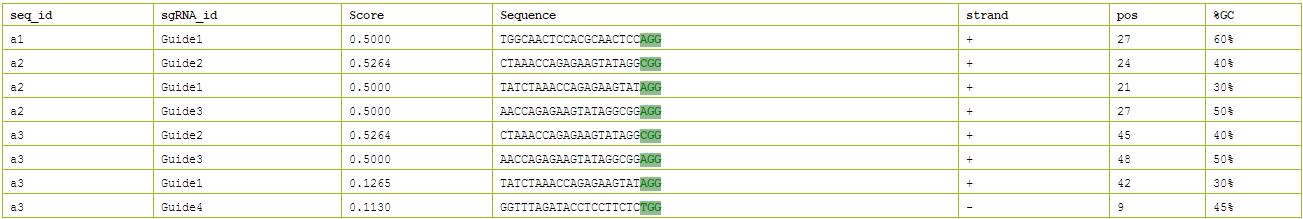
In page of "design" user can custom sgRNA multi-sequences in FASTA that more than 30 nt (included PAM "NGG"): Custom sgRNA sequence in format "NNNN + spacer (20bp) + NGG + NNN" or long sequences containing one or more sgRNA sequence of the format. Input content exampled in text as followed, the result of on-target score of target sequences are list in Figure 6.
GTAATAATGGCAACTCCACGCAACTCCAGG
>a2
GTATCTAAACCAGAGAAGTATAGGCGGAGG
>a3
GTATCTAAACCAGAGAAGGAGGTATCTAAACCAGAGAAGTATAGGCGGAGG
5. Design of Score System
Design of On-target Score and Off-target Score system.
- 5.1 Score for sgRNA effective
- 5.2 Score for off-target potential
![]()
![]()
The Nucleotide preference and GC conetent of sgRNA are concidered to score for the sgRNA effective. The score range from 0 to 1, the high score presents the high effective of sgRNA.
![]()
In the off-target score model, sgNRA mismatche's possition, number and identity, and PAM's mismatches are taken into account. The score range from 0 to 1, site with higher off-target score has much off-target potential, which ususally should be avoided.
In general, sgRNA with less high off-target score sites and with high on-target score are the ideal ones user needs.
6. Contact
Project Leader: Lingling Chen, Kabin Xie
Any question contact us
