CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is an acronym for DNA loci that contain multiple, short, direct repetitions of base sequences. Each repetition contains a series of base pairs followed by the same or a similar series in reverse and then by 30 or so base pairs known as "spacer DNA". CRISPRs are found in the genomes of approximately 40% of sequenced bacteria and 90% of sequenced archaea (Grissa et al., 2007). CRISPRs are often associated with cas genes which code for proteins that perform various functions related to CRISPRs. The CRISPR-Cas system functions as a prokaryotic immune system, in that it confers resistance to exogenous genetic elements such as plasmids and phages and provides a form of acquired immunity. CRISPR spacers recognize and silence exogenous genetic elements in a manner analogous to RNAi in eukaryotic organisms (Marraffini and Sontheimer, 2010).
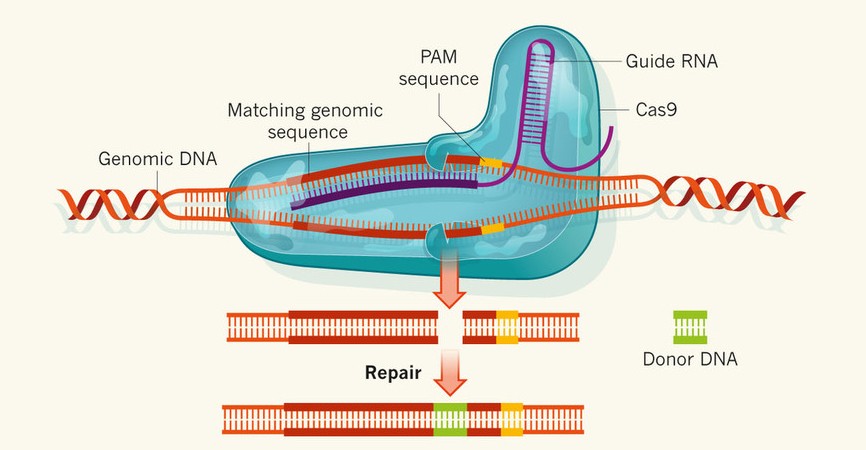
We develop a web application tool, CRISPR-P, for CRISPR single-guide RNA (sgRNA) design in many plant species. CRISPR-P allows users to search for high specificity Cas9 target sites within DNA sequences of interest, which also provides off-target loci prediction for specificity analyses and marks restriction enzyme cutting site to every sgRNA for further convenient in experiment.
Reference
Yang Lei#, Li Lu#, Hai-Yang Liu, Sen L, Feng Xing, Ling-Ling Chen*. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant, 2014, 7(9): 1494-1496. doi: 10.1093/mp/ssu044.
